Contributors
Rafael Pineda-Alemana, Antistio Alviz-Amadorb, Rodrigo Galindo-Murilloc, Humberto Perez-Gonzalezd, Erika Rodriguez-Cavallob, Dario Mendez-Cuadro*e
a) Analytical Chemistry and Biomedicine Group, Medicine Faculty, University of Cartagena, Cartagena, Colombia
b) Analytical Chemistry and Biomedicine Group, Pharmaceutical Sciences Faculty, University of Cartagena, Cartagena, Colombia
c) Department of Medicinal Chemistry, University of Utah, Salt Lake City, UT, USA
d) Department of Mathematics, Exact and Natural Sciences Faculty, University of Cartagena, Cartagena, Colombia
e) Analytical Chemistry and Biomedicine Group, Exact and Natural Sciences Faculty, University of Cartagena, Colombia
Reference
Pineda-Aleman R, Alviz-Amador A, Galindo-Murillo R, Perez-Gonzalez H, Rodriguez-Cavallo E, Mendez-Cuadro D.
J Mol Graph Model. 2023, 124, 108533
doi: 10.1016/j.jmgm.2023.108533. Epub ahead of print. PMID: 37311331.
Cysteine carbonylation with reactive carbonyl species from lipid peroxidation induce local structural changes on thioredoxin active site.
Cysteine carbonylation with reactive carbonyl species from lipid peroxidation induce local structural changes on thioredoxin active site
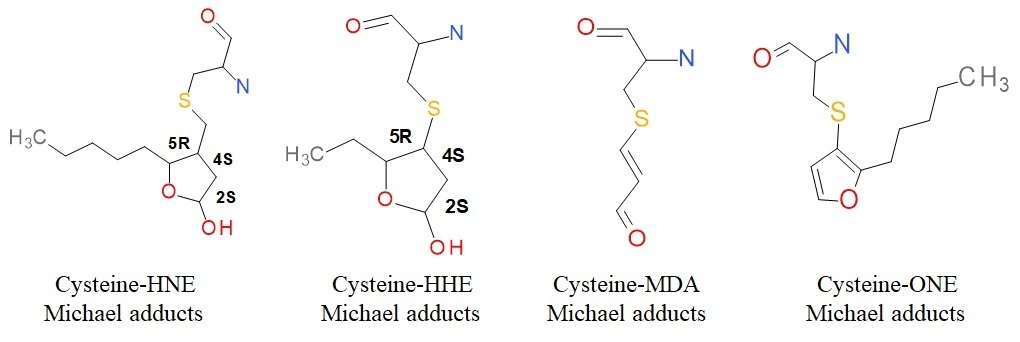
Protein oxidative modifications with reactive carbonyl species (RCS) is directly linked to metabolic processes in premature aging, cancer, neurodegenerative and infectious diseases. RCS as 4-Hydroxy-2-nonal (HNE), 4-Hydroxy-2-hexenal (HHE), 4-Oxo-2-nonenal (ONE) and Malondialdehyde (MDA) attack nucleophilic amino acids residues forming irreversible adducts with proteins as Thioredoxins (Trx). This is a class of small thiol oxidereductases playing a key role in redox signaling and oxidative stress responses in mammals. Although proteomic studies have identified to Cys-32 residue as a target of HNE attack that inhibit its enzymatic activity, how this carbonylation affects its structure and dynamic behavior at the atomic level is unknown. Even more, the molecular bases for the atomistic behavior of these modified proteins have not been completely understood. We present molecular dynamics simulations of Trx-modified with four different RCS to analyze its global and local structural effects. For this, parameters supported in the AMBER force fields were built and validated for three non-natural cysteine residues modified with HHE, ONE and MDA. Results obtained showed a slight change in the global conformational stability of Trx modified with HNE and MDA, establishing that all modified proteins presented local regions of high mobility in the modified catalytic site and some regions far from the modification area. In addition, essential domain movement modes evidences that proteins modified with the RCS assayed induce changes in conformational flexibility. Finally, these data showed that the given conformational changes did not cause global changes in proteins but rather localized changes in particular regions.
Comments
Design of new non-standard cysteine residues with lipid peroxidation derivatives
The new parameters of modified lysine with HHE, MDA, and ONE were created by MSc Rafael Pineda-Aleman (rpinedaa@unicartagena.edu.co), with the advice and support of professors Humberto Perez-Gonzalez (hperezg@unicartagena.edu.co), Rodrigo Galindo-Murillo (rodrigo.galindo@utah.edu), Ricardo Vivas-Reyes (rvivasr@unicartagena.edu.co), and Dario Mendez-Cuadro (dmendezc@unicartagena.edu.co).
Parametrization of the three new modified amino acid residues (Cys-HHE, Cys-MDA, and Cys-ONE) was developed using the AmberTools2016 software package. Analogous parameter generation was performed using the antechamber programs with the General Amber Force Field (GAFF), parmcal, and parmchk. The parameters and typology were created in the Leap program using the tleap version (without a graphical interface) and xleap (with graphical interface).
Step 1. Design non-standard residue cysteine with HHE, MDA and ONE were generated using the Gauss View Software version 5.
Step 2. Geometric optimization of the amino acid structure was carried out considering the theoretical level: #HF/6-31G* SCF=tight test Pop=MK iop (6/33=2) iop(6/42=6) opt.
Step 3. Construction of the parameter file, file.frcmod:
> antechamber -i init-gau.log -fi gout -o u01.prepin -fo prepi -c resp -s 2 -rn U01 -at gaff2 -nc 0
> parmchk -i u01.prepin -f prepi -o u01.frcmod
Step 4. Construction of the library file, file.lib, using the Leap program:
> source leaprc.gaff2
> loadamberparams u00.frcmod
> loadamberprep u00.prepin
> Desc U01
> Saveoff U00 u00.lib
[ APD Home ] [ Submit Parameters ] [ Amber Home ] [ UK Mirror ] [ Pharmacy Home ] [ Contact ]
Last modified
17 Sep 2012

